Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
C: ATP AND OXIDATIVE PHOSPHORYLATION
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8C: After class and this reading, students will be able to
|
C10. Proton Gradient Collapse and ATP synthesis - Structure
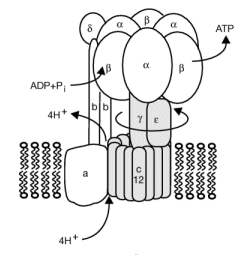
The mechanism by which the proton gradient drives ATP synthesis involves a complex coupling of the F0 and F1 subunits. A more detailed image of the whole ATO synthase complex is shown below.
Figure: Detailed View of F0F1 ATP synthase structure
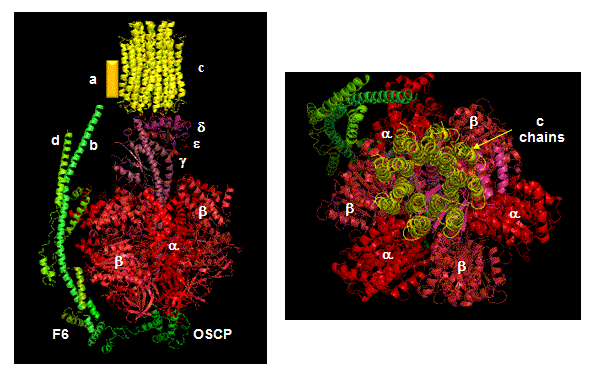
Closer views of the c subunits and a yellow rectangle representing the a subunit (missing in the combined crystal structure) comprise the Fo part of the complex are shown below. These subunits reside in the inner membrane of the mitochondria (or cell membrane of bacteria) and are involved in proton transport from matrix (or cytoplasm of a bacteria) to the inner membrane space (or periplasmic space of bacteria). The multiple c subunits consist of two very hydrophobic helices connected by a loop in a helix-loop-helix motif.
![]() 7/127/17: The following Jmol links contains multiple views of the
FoF1ATPase. It is repeated several
times below.
7/127/17: The following Jmol links contains multiple views of the
FoF1ATPase. It is repeated several
times below.
![]() Jmol:
Bovine F0F1 ATPase
aka ATP Synthase (under development)
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Bovine F0F1 ATPase
aka ATP Synthase (under development)
Jmol14 (Java) |
JSMol (HTML5)
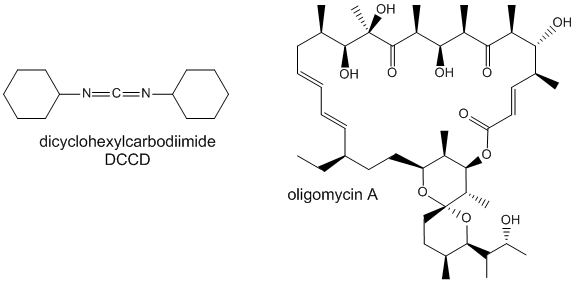
Two classic inhibitors (structures shown below) of ATP synthase interact with the Fo subunit. One, oligomycin A, binds between the a and c subunits and blocks proton transport activity of the Fo subunit. Oligomycin A sensitivity requires, paradoxically, OSCP (Oligomycin-Sensitivity Conferring Protein which is analogous to the bacterial delta subunit), a stalk protein subunit distal to Fo which couples Fo and F1. Another inhibitor, dicyclohexylcarbodiimide reacts with a protonated Asp 61 in c subunits of F0. It does so even at pH 8.0 which indicates that the pKa of the Asp 61 is much higher than usual. This might occur if the Asp is a very hydrophobic environment. The modification of one As 61 in only one c subunit is necessary to stop Fo activity. The protonated carboxyl group donates a proton to a N atom in DCCD, which then reacts with the deprotonated Asp to form an O-acyl isourea derivative.
Figure: Structure of Oligomycin A and DCCD - Inhibitors of proton transport by Fo
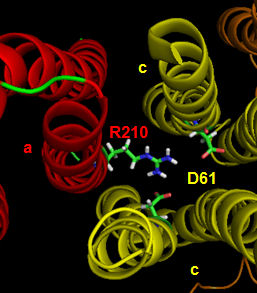
The figures below shown the structure of the ac complex from E. Coli. Protons flow to the a chain Arg 210 which is between two Asp 61 on adjacent c chains. One of the Asp 61 is protonated allowing it to alter conformation and essentially rachet in the membrane domain in a motional faciliated by the development of a neutral protonated Asp.
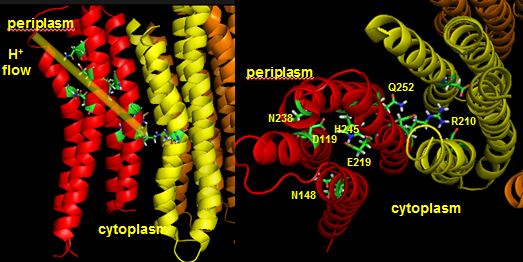
Protons from the inner membrane space or in the periplasmic space (in the above figures) then flow from the periplasm by forming a "handshaking" proton transfer relay which delivers another proton to the deprotonated Arg 210 allowing the circular ratching of the c subunits in the membrane to continue. A set of polar residues entirely within subunit a, including Gln 252, Asn 214, Asn 148, Asp 119, His 245, Glu 219, Ser 144 and Asn 238 provide the path as illustrated below.
When a proton is passed to the unprotonated Asp 61, a conformational change in the protonated c subunit occurs. This leads to changes in c subunit interactions which seems to ratchet the c12 core. Since the c12 oligomer contacts the γsubunit connecting the Fo stalk and F1 ATPase units, the γ subunit rotates, leading to sequential conformational changes in each of the 3 contacted (αβ)2 dimers of the F1 enzyme. This leads to changes in ATP affinity through cycling each through the L, O, and T conformations.
Figure: Coupling Proton Flow in F0 to Conformation Change
Reprinted by permission of Nature. Rastogi & Girvin. Nature 402, 263-268 (1999) Copyright 1999 McMilllan Publishers LTD
![]() animation 2: ATP synthesis mechanism
animation 2: ATP synthesis mechanism
![]() Molecular
Movies (scroll down to ATP synthase)
Molecular
Movies (scroll down to ATP synthase)
![]() Mitochondria:
Harvard
Mitochondria:
Harvard
![]() Powering the
Cell: Mitochondria
Powering the
Cell: Mitochondria
![]() XVIVO: Scientific Animations
XVIVO: Scientific Animations
![]() ATPase
animations from the MRC
ATPase
animations from the MRC
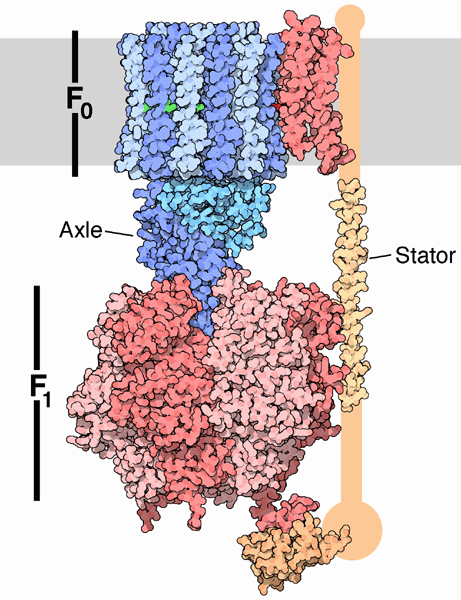
In summary, FOF1ATPase (or synthase) is a rotary enzyme that ultimately couples collapse of a proton gradient (a chemical potential gradient which contributes to the transmembrane electrical potential) to a chemical (phosphorylation) step. The rotor, which is in contact with both the FO proton pore, and the F1 synthase, moves with respect to both subunits, which couples them. Motion of course is relative so the rotor can be thought of as static with the FO and F1 subunits as rotating. The FO pore can hence to be considered an electrical motor and the F1 synthase a chemical motor. Carrying the analogy of a motor even further, the FO electrical motor turns the F1 chemical motor into a generator, not of electricity but of ATP. The figure and link below, taken from the Protein Data Bank, go into more depth about this nanomotor.
Navigation
Return to Chapter 8C: ATP and Oxidative Phosphorylation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8C: ATP and Oxidative Phosphorylation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.