Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
C: ATP AND OXIDATIVE PHOSPHORYLATION
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8C: After class and this reading, students will be able to
|
C9. ATP synthase
ATP synthase, also called FoF1ATPase, is a rotary motor enzyme. This enzyme is found in the inner membrane of mitochondria, the analogous thylakoid membranes of chloroplasts, and in the cell membrane of bacteria. The enzyme consists of two parts, the membrane bound Fo which is a proton translocator, and the F1 part which has catalytic activity. The F0 part can be considered to be a rotary electrical motor powered by proton flow, while the F1 part acts as a rotary chemical motor powered (in reverse) by ATP hydrolysis. The enzyme is reversible. If protons flow down a concentration gradient through Fo, ATP is synthesized by F1. Alternatively, ATP hydrolysis by F1 leads to transport of protons through Fo and against a concentration gradient. Isolated F1 can only break down ATP, and not synthesize it.
![]() Summer 2017: The following Jmol links contains multiple views of the complex. It is repeated below.
Summer 2017: The following Jmol links contains multiple views of the complex. It is repeated below.
![]() Jmol: FoF1
ATPase aka ATP Synthase
Jmol14 (Java) |
JSMol (HTML5)
Jmol: FoF1
ATPase aka ATP Synthase
Jmol14 (Java) |
JSMol (HTML5)
The F1 subunit (with quaternary structure of a3b3 forming a hexagonal ringed structure with a central cavity,occupied by a gamma subunit)is about 80 angstroms from the Fo subunit and both are connected to the rod-shaped γ subunit which spans the center of the a3b3 ring. Energy transduction (necessary to capture the negative free energy change associated with the collapse of the proton gradient to drive the positive free energy change for ATP synthesis) occurs between the two subunits. Noji investigated the structural changes in the γ subunit, wishing to get direct experimental evidence for Boyer's three-state conformational model (L-O-T) for ATP synthesis.
Figure: Boyers three-state conformational model (L-O-T) for ATP synthesis
In this model, the F1 subunit exists in three states: an O - open - state with very low affinity for substrates and has no catalytic activity; a L - loose - state with low affinity for substrates and also no catalytic activity, and a T - tight - tight state with high affinity for substrates and with catalytic activity. The F1 subunit consist of three α and three β subunits, which can cycle between three conformations, bind substrate, and have catalytic activity. The collapse of the proton gradient (i.e. the proton-motive force) causes the γ subunit to rotate like a crankshaft relative to the F1 subunit, forcing the β subunit to change conformation from the T to the O (releasing ATP) and then the L (binding ADP and Pi). The γ subunit does not appear to undergo any significant conformational change on ATP hydrolysis as evidenced by tritium exchange studies of amide protons. To prove that the γ subunit rotates, you'd have to observe a single molecule. Since the γ subunit was too small to visually discern its rotation, Noji covalently attached a fluorescein-labeled protein filament called actin to the γ subunit (near where Fo would bind). He then fixed the whole F1 molecule to a glass slip through the a3b3 part, immobilizing that part of the molecule. The γ subunit was free to rotate, which could be detected by observing the fluorescence under a fluorescent microscope from the attached actin filament.
Figure: fluorescein-labeled protein filament called actin to the γ subunit
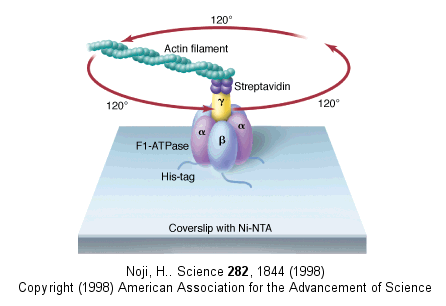
The actin filament rotated only in the presence of ATP. It rotated only counterclockwise, and continued for 10 minutes. This demonstrated that the motion was not random, but a specific motion of the γ subunit. At extremely low concentration of ATP, rotation occurred only in 120o increments, implying one step per molecule of ATP hydrolyzed. (Remember the β subunits are separated by 120o ). As the rotation occurs, there is viscous resistance to movement of the actin filament. He calculate that for a single 120o step caused by hydrolysis of a single ATP molecule, the amount of work was 80 piconewton which is about the free energy of hydrolysis of a single ATP molecule. Incidentally, Boyer was awarded the Noble prize in Chemistry in 1997 for his work. Noji and his colleagues (Nature, 410, 898 (2001), replaced the actin filament with a smaller colloidal gold bead (40 nm diameter) with less frictional resistance to movement and used laser light scattering to probe the rotation of the fixed F1 subunit through the γsubunit.
Figure: smaller colloidal gold bead (40 nm diameter) with less frictional resistance
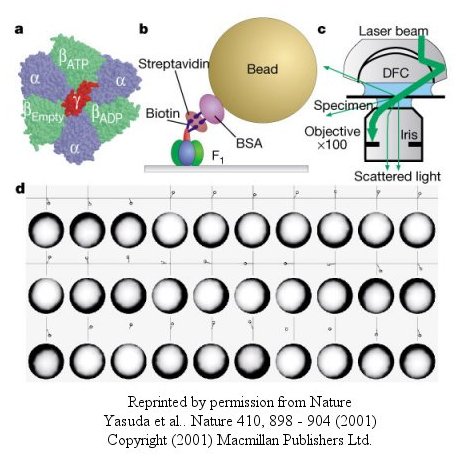
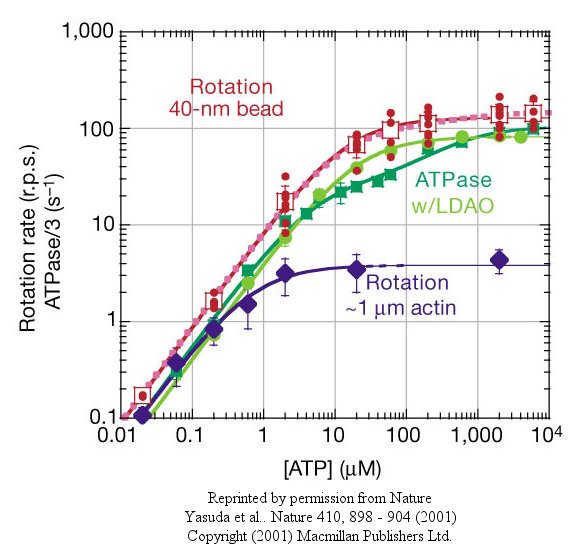
At low [ATP], the motor rotates in 120o steps. At high [ATP], the rotation rate becomes continuous and saturates (with Michaelis/Menten kinetics) at 130 revolutions per second.
Figure: rotation rate becomes continuous and saturates
Recent experiments (Wantanbe, 2010) using immobilized ATPase and magnetic tweezers have addressed the timing of substrate binding and product release when the enzyme is run in reverse (ATP hydrolysis). On rotation of the gamma subunit, the three binding sites change properties. In hydrolysis, ATP binds to the open site, and helps promote the 120 degree rotation. In the next step, ATP is hydrolyzed. In final step, products dissociate. Pi dissociation occurs last from the third site. Hence each of the 3 beta binding sites have different roles. One binds substrate, one performs catalysis, and third releases products. Assuming the synthesis pathway is the reverse of the ATPase reaction, the final release of Pi in ATP cleavage predicts that Pi binds first in the synthetic direction. This would preclude the binding of ATP next which is critical since its concentration during synthesis can be 10x higher than that of ADP. As Pi is bound first, only ADP, not ATP can bind next.
The gamma subunit rotation plays a "catalytic" role as its rotation induces cyclic conformational changes in the beta subunit of the synthase. Can ATP synthesis occur without the gamma subunit by a mechanisms which involves a less proficient, but concerted set of cyclic change in beta subunit conformation? Apparently it can. Uchihashi et al have used high speed atomic force microscopy (AFM) to study the (alpha-beta)3 hexagon from the F1 subunit without the gamma subunit. They found that upon ATP hydrolysis, the beta subunits underwent conformational changes in the same counterclockwise rotary direction as when the gamma subunit was present.
Figure: AFM Study of Conformational Changes in F1 "gammaless" subunit
These experiments conclusively show that the F1 subunit is effectively a rotary motor with the gamma subunit acting as a rotor in the stationary hexagon ring composed of the 3 pairs of alpha/beta subunits which acts as the stator (stationary part of an electric rotary motor).
http://www.powerpulse.net/techPaper.php?paperID=142&page=3
![]() Jmol: FoF1
ATPase aka ATP Synthase
Jmol14 (Java) |
JSMol (HTML5)
Jmol: FoF1
ATPase aka ATP Synthase
Jmol14 (Java) |
JSMol (HTML5)
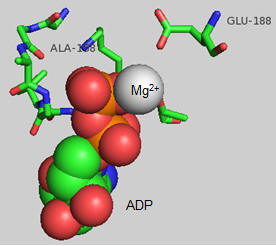
The actual amino acids involved in the mechanism of ATP synthesis/hydrolysis are still not clearly defined but Glu 190 on the beta subunit clearly acts as a general base. The figure below shows bound ADP and the proximity of Glu 188. Ala 158 is thought to move towards the active site after a conformational change, with the nonpolar methyl side chain displacing an adjacent water molecule which could leave as a product of ATP synthesis.
Navigation
Return to Chapter 8C: ATP and Oxidative Phosphorylation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8C: ATP and Oxidative Phosphorylation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.