Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 9 - SIGNAL TRANSDUCTION
D: mTOR and Nutrient Signaling
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 6/22/16
|
Learning Goals/Objectives for Chapter 9D:
|
D2. mTOR Inhibition by Rapamycin
The structure of the mTORC1 complex is complex itself, in part, since some of its components were studied and named before their roles in the mTORC1 complex were elucidated. Investigators were obviously interested in the molecular target(s) of rapamycin. Yeast (a fungus) was an easy organism to study using genetic techniques. Three genes were found that when mutated inhibited the effect of the inhibitor rapamycin (i.e. so that rapamycin did not inhibit mTOR). Two of these were the Ser/Thr kinases mTOR1 and mTOR2. The other was an analog of a protein found in humans (FK Binding Protein 12 - FKBP12). The family of FK binding proteins act as protein chaperones and have Pro-X peptidyl prolyl isomerase activity (PPI). FKBP12 specifically binds a drug (FK506 also called Tacrolimus) that is an immunosuppressant (do you see the general link with immune cell division and growth?). The binary complex of FK506 and FKBP12 bind a third protein, a phosphatase called calcineurin, and block its phosphatase activity and signal transduction required for T cell activation and proliferation. This activity of FKBP12 strangely does not require its PPI activity.
Now back to mTORC1. Here are the known components of the core complex:
- mTOR, a Ser/Thr kinase;
- Raptor, a Regulatory-associated protein of mTOR; it modulates the specificity of the kinase;
- mLST8, the mammalian lethal with SEC13 protein 8 (also called Mammalian Lethal With SEC13 Protein 8 e
In addition, other proteins associate with the core complex
- PRAS40, a proline-rich Akt substrate of Akt
- DEPTOR , DEP Domain Containing MTOR-Interacting Protein (where DEP is Dishevelled, Egl-10 and Pleckstrin domain found in these 3 proteins and others involved in G-protein signaling)
- FKBP, which binds rapamycin.
mTORC2 is not sensitive to rapamycin. Instead of RAPTOR, it has a protein called RICTOR (rapamycin insensitive companion of TOR).
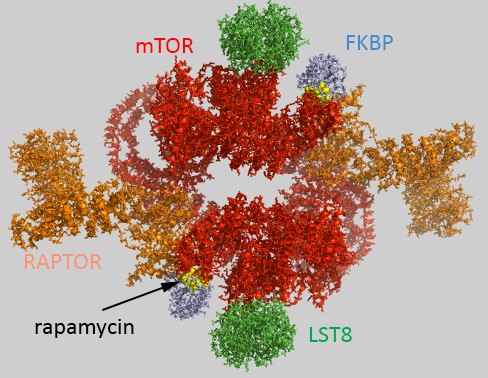
A structure (5flc) of the human mTORC1 containing mTOR, Raptor and mLST8 bound to FK506 binding protein (FKBP)-rapamycin complex has been solved with cryomicroscopy. An image created by Pymol and a 3D Jsmol structure are shown below.,
![]() Jmol:
human mTORC1 (5flc)
Jmol14
(Java) |
JSMol (HTML5)
Jmol:
human mTORC1 (5flc)
Jmol14
(Java) |
JSMol (HTML5)
The complex acts physiologically as a dimer with rotational C2 symmetry.
- chains B,F ,1-4: pieces of mTOR (mol ID 1-3) as homodimer - Red;
- chains A and E: Raptor (mol id 4) orange;
- chains D and H: LST8 (mol id 5) green;
- chains C and G: FKBP (mol id 6) light blue;
- rapamycin in C and G, yellow spacefill.
Raptor has been liken to tape as it interacts with the two mTOR subunits, holding them together into a larger, donut-like structure, stabilizing the dimer.
How does rapamycin inhibit mTOR? You have studied many kinds of inhibitors (competitive, uncompetitive, mixed or noncompetitive inhibition) in which the inhibitor binds to either free E or the ES complex. The figure above suggests that rampamycin binds at the interface of the mTOR kinase (red) and FKBP. Presumably the RAP:FKBP complex binds to mTOR.
The figure below shows how rapamycin (shown in sticks) is sandwiched between mTOR (red) and FKBP (light blue). This offers clues as to how it inhibits mTOR. The rapamycin:FKBP complex is an allosteric inhibitor with its effect dependent on both substrate and post-translational phosphorylation.
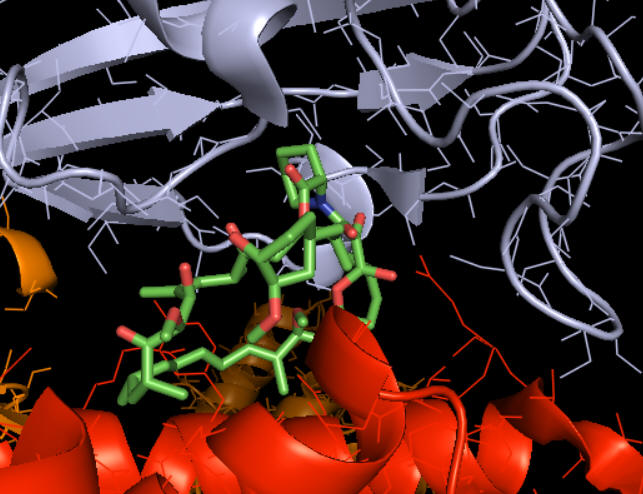
Right across from the light blue FKBP protein is the green LST8 protein (see above). In between these is a cleft which is the active site. One could imagine that RAP:FKBP binding to mTOR might interfere with substrate binding in mTORC1 but not in mTORC2. RAPTOR in mTORC1 probably helps recruit substrates and hence is involved in determination of substrate specificity. RAPTOR interacts with short section called the TOR signaling (TOS) motif in mTORC1 substrates. The part of RAPTOR that binds TOS in substrates is at the base of the mTORC1 active site, probably narrowing it further as it provides exquisite substrate selectivity.
Navigation
Return to Chapter 9D. Nutrient Signaling Sections
Return to Biochemistry Online Table of Contents

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.