Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
C: ATP AND OXIDATIVE PHOSPHORYLATION
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8C: After class and this reading, students will be able to
|
C6. Complex III
![]()
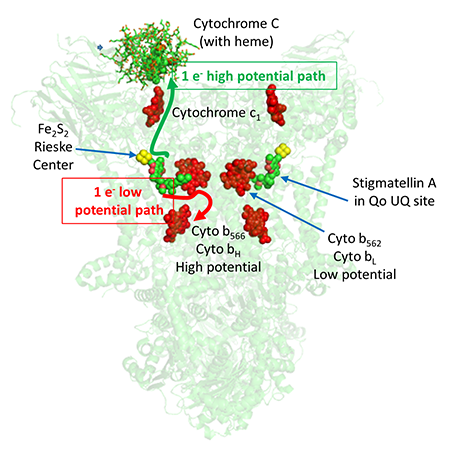
Complex III is a complicated, multisubunit protein. The subunits involved in electron transfer are cytochrome b, cytochrome c1 and the Rieske iron sulfur protein (ISP). Cytochrome b has two hemes. One is cyto b562 which is also called the low potential heme or cyto bL. The other is cyto b566 which is called the high potential heme or cyto bH. The cytochrome c1 subunit has one heme.
The following Jmol links contains multiple views of the complex. It is repeated several times below.
![]() Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
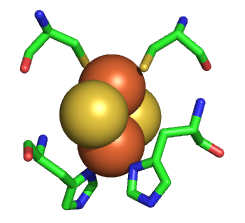
The Rieske iron sulfur protein has a Fe2S2 iron sulfur cluster which differs from other such clusters in that each Fe is also coordinated to two His side changes, as shown in the figure below. Alterations in H bonds to the histidines and to the sulfurs in the complex can dramatically affect the standard reduction potential of the cluster.
![]() Jmol:
Rieske Center of Complex III
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Rieske Center of Complex III
Jmol14 (Java) |
JSMol (HTML5)
As with complex I and IV, proton and electron transfer are coupled processes. However, in contrast to Complex I, in which protons pass through protein domains that have homology to K+/H+ antiporters, and Complex IV, in which they pass through a combination of a water channel and the H-bond network, the protons in Complex III are carried across the inner membrane by ubiquinone itself. Two reduced ubiquinones (UQH2) from complex I pass their four matrix-derived protons into the inner membrane space. In the process four electrons are removed in a multiple step process called the Q cycle.
The two electrons from each UQH2 take different paths. One electron moves to a Fe/S Rieske cluster and the other to cytochrome bL. The electrons moved to the Rieske center then moves to cytochrome c1s and then to the mobile electron carrier cytochrome C which is bound to the complex in the intermolecular space. The electrons moved to cyto bLs are transferred to cytochrome bH in the complex. Though this latter path, two electrons (from two UQH2) are then moved to oxidized UQ, and two matrix protons are added to reform one UQH2. Hence, only one UQH2 participates in the net reaction shown as below.
QH2 + 2 cyto c3+ + 2H+matrix → Q + 2 cyto c 2+ + 4H+IMS
This net overall reaction, the Q cycle, is illustrated below. This net overall reaction, the Q cycle, is illustrated below.
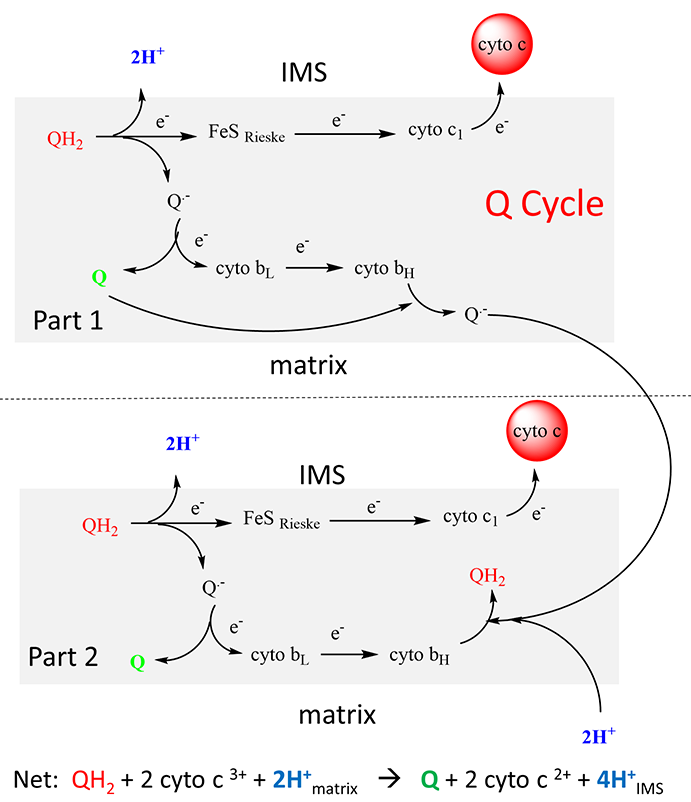
Once again, there are no “proton” channels or H bonded networks in the protein for proton transfer across the inner membrane.
The figure below shows the relative position of the bound mobile electron carrier, cytochrome C, and the internal ones, the Rieske Fe/S cluster and cytochrome bL and bH. Note also the molecule stigmatellin A, which binds to the site where UQ becomes reduced (called the Qo site) and inhibits the complex. This shows that UQ/UQH2 are in position to react readily with the Rieske canter and cytochrome bL heme.
![]() Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
Another way to think about the electron transfer process from UQH2 to cytochrome C is that the 2 electrons from UQH2 take two different paths, one a high potential path to the Rieske center and on to cytochrome C, and another low potential path to the bL heme and on to the bH heme and then to UQ to reform UQH2 (see figure above).
Complex III, along with Complex I, can also produce unwanted reactive oxygen species (ROS). Only three of the protein subunits, cytochrome b (with the bL and bH hemes), cytochrome c1, and the Rieske iron sulfur protein (ISP) are involved in electron transfer, so one of those is mostly likely involved in ROS production. Experiments and mathematical models support a mechanism that involves a reduction of UQ by addition of one electron from cytochrome bL to form UQ. which then passes its electron on to dioxygen to form superoxide (O2-.).
As two ubiquinones must bind to the complex, there must be two proximal sites. One is the Qi site where oxidized UQ binds and receive an electron. The other is the Qo site where UQH2 binds.
From a kinetic perspective, the first UQH2 binds and transfers two electrons, one to the Rieske cluster (and on to cytochrome c1 and then to cytochrome C) and one to cytochrome bL (and on to heme bH) and then to an oxidized UQ bound at the Qi site. The UQ. radical is stabilized by the adjacent bH heme which has a lower affinity for electrons. Now a second UQH2 binds to the Qo site, and transfers two electrons, again one via the Rieske cluster and the second through cytochrome bL and bH to the UQ. radical present at the Qi site to form UQH2 after two protons are transferred to it from the matrix.
Now a second UQH2 binds to the Qo site, and transfers two electrons, again one via the Rieske cluster and the second through cytochrome bL and bH to the UQ. radical present at the Qi site to form UQH2 after two protons are transferred to it from the matrix.
Antimycin A, an extremely toxic drug, binds to the UQ Qi site and hence blocks electron transfer from cytochrome bL to bH at the Qi site. Heme bL can then pass its electron to dioxygen to produce superoxide.
![]() Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Complex III
Jmol14 (Java) |
JSMol (HTML5)
Navigation
Return to Chapter 8C: ATP and Oxidative Phosphorylation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8C: ATP and Oxidative Phosphorylation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.