Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
C: UNDERSTANDING PROTEIN CONFORMATION
BIOCHEMISTRY - DR. JAKUBOWSKI
3/4/16
|
Learning Goals/Objectives for Chapter 2C: After class and this reading, students will be able to
|
C4. Common Motifs in Proteins
Super-Secondary Structure - Given the number of possible combinations of 1o, 2o, and 3o structures, one might guess that the 3D structure of each protein is quite distinctive. This is true. However, it has been found that similar substructures are found in proteins. For instance, common secondary structures are often grouped together to form a motifs called super-secondary structure (SSS). See some examples below:
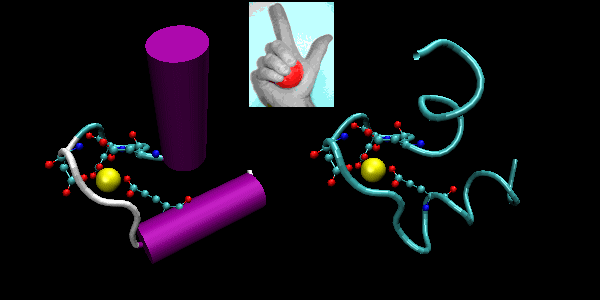
- helix-loop-helix : found in DNA binding proteins and also in calcium binding proteins. This motif, which is also a helix-loop-helix, is often called the EF hand. The loop region in calcium binding proteins are enriched in Asp, Glu, Ser, and Thr. Why? The EF hand shown below is from calmodulin.
Figure: helix-loop-helix (image made with VMD)
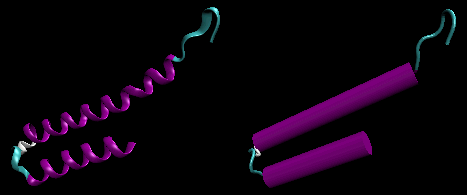
Figure: EF Hand
![]() Jmol:
Updated helix-loop-helix of the lambda Repressor
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated helix-loop-helix of the lambda Repressor
Jmol14 (Java) |
JSMol (HTML5)
![]() Jmol:
Updated helix-loop-helix (EF hand) from calmodulin
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated helix-loop-helix (EF hand) from calmodulin
Jmol14 (Java) |
JSMol (HTML5)
- beta-hairpin or beta-beta: is present in most antiparallel beta structures both as an isolated ribbon and as part of beta sheets.
Figure: beta-hairpin, or beta-beta (image made with VMD)

![]() Jmol:
Updated beta-hairpin from bovine pancreatic trypsin inhibitor
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated beta-hairpin from bovine pancreatic trypsin inhibitor
Jmol14 (Java) |
JSMol (HTML5)
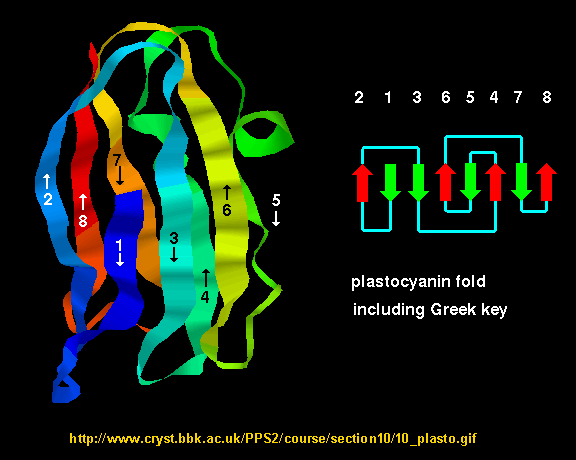
- Greek Key motif: four adjacent antiparallel beta strands are often arranged in a pattern similar to the repeating unit of one of the ornamental patterns used in ancient Greece.
Figure: Greek Key Motiff
![]() Jmol:
Greek Key
Jmol:
Greek Key
- Figure: beta-alpha-beta: is a common way to connect two parallel beta strands. (beta hairpin used for antiparallel beta strands).
Figure: beta-alpha-beta (image made with VMD with H atoms added by Molprobity
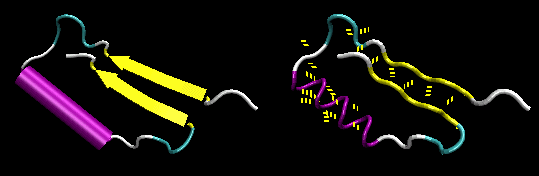
![]() Jmol: Updated
beta-helix-beta motif from triose phosphate isomerase
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
beta-helix-beta motif from triose phosphate isomerase
Jmol14 (Java) |
JSMol (HTML5)
-
Beta Helices: These right-handed parallel helix structures consists of a contiguous polypeptide chain with three parallel beta strands separated by three turns forming a single rung of a larger helical structure which in total might contain as many as nine rungs. The intrastrand H-bonds are between parallel beta strands in separate rungs. These seem to prevalent in pathogens (bacteria, viruses, toxins) proteins that facilitate binding of the pathogen to a host cell.
Figure: Beta Helices (image made with VMD)
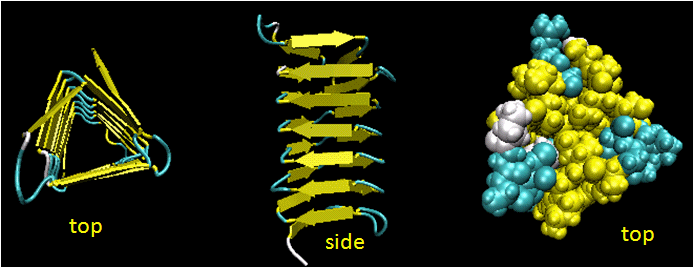
Table: Beta Helices
Vibrio cholerae
cholera
Helicobacter pylori
ulcers
Plasmodium falciparum
malaria
Chlamyidia trachomatis
VD
Chlamydophilia pneumoniae
respiratory infection
Trypanosoma brucei
sleeping sickness
Borrelia burgdorferi
Lymes disease
Bordetella parapertussis
whooping cough
Bacillus anthracis
anthrax
Neisseria meningitides
menigitis
Legionaella pneumophilia
Legionaire�s disease
-
 of
the Swiss Institute of Bioinformatics.
(SIB) is dedicated to the analysis of protein sequences and structures
as well as 2-D PAGE
of
the Swiss Institute of Bioinformatics.
(SIB) is dedicated to the analysis of protein sequences and structures
as well as 2-D PAGE
Domains -
Domains are the fundamental unit of 3o structure. It can be considered a chain or part of a chain that can independently fold into a stable tertiary structure. Domains are units of structure but can also be units of function. Some proteins can be cleaved at a single peptide bonds to form two fragments. Often, these can fold independently of each other, and sometimes each unit retains an activity that was present in the uncleaved protein. Sometimes binding sites on the proteins are found in the interface between the structural domains. Many proteins seem to share functional and structure domains, suggesting that the DNA of each shared domain might have arisen from duplication of a primordial gene with a particular structure and function.
Evolution has led towards increasing complexity which has required proteins of new structure and function. Increased and different functionalities in proteins have been obtained with additions of domains to base protein. Chothia (2003) has defined domain in an evolutionary and genetic sense as "an evolutionary unit whose coding sequence can be duplicated and/or undergo recombination". Proteins range from small with a single domain (typically from 100-250 amino acids) to large with many domains. From recent analyzes of genomes, new protein functionalities appear to arise from addition or exchange of other domains which, according to Chothia, result from
- "duplication of sequences that code for one or more domains
- divergence of duplicated sequences by mutations, deletions, and insertions that produce modified structures that may have useful new properties to be selected
- recombination of genes that result in novel arrangement of domains."
Structural analyses show that about half of all protein coding sequences in genomes are homologous to other known protein structures. There appears to be about 750 different families of domains (i.e small proteins derived from a common ancestor) in vertebrates, each with about 50 homologous structures. About 430 of these domain families are found in all the genomes that have been solved.
Navigation
Return to Chapter 2C: Understanding Protein Conformation Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2C: Understanding Protein Conformation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.