Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 6 - TRANSPORT AND KINETICS
A: PASSIVE AND FACILITATED DIFFUSION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 04/08/16
|
Learning Goals/Objectives for Chapter 6A: After class and this reading, students will be able to
|
A3. "Receptors" in Facilitated Diffusion
Two types of proteins are involved in facilitated diffusion. Carrier proteins (also called permeases or transporters) such as the glucose transporter (GLUT1) move solute molecules across a membrane, Channels facilitate diffusion of ions down a concentration gradient. In the former cases, the ligand binds the receptor (permease, transport protein) which induces a conformational change in the receptor as illustrated in the animation above and in the figure link below.)
Figure: Models for facilitated diffusion of glucose
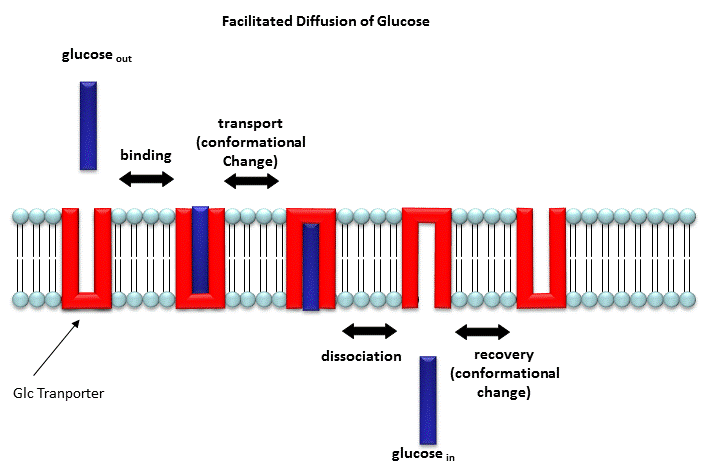
In the latter case, a ligand can bind to the receptor (channel protein) which induces a conformational change in the receptor, a "ligand-gated" channel through the membrane. This process would lead to the diffusion of many ions across the membrane (down a concentration gradient) until the channel closes (which can be induced by ligand dissociation or other events). Clearly, the mathematics we derived for the carrier proteins does not apply to the channel proteins. In addition, there are other ways to "gate" open a channel protein, which we will discuss later. Also some transporters can move solute molecules across a membrane against a concentration gradient. These proteins require an external energy source (like ATP or coupling to the favorable collapse of a second transmembrane gradient ) to drive this thermodynamically unfavored process.
Both links above are from the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. These molecular dynamic simulations were made with VMD/NAMD/BioCoRE/JMV/other software support developed by the Group with NIH support.
Recently, the x-ray structures of two transporters that are powered by the collapse of a second gradient, were reported.
![]() Jmol:
Updated
Lactose Permease
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated
Lactose Permease
Jmol14 (Java) |
JSMol (HTML5)
![]() Jmol:
Updated Glycerol-3-Phosphate Transporter
From E. Coli
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Glycerol-3-Phosphate Transporter
From E. Coli
Jmol14 (Java) |
JSMol (HTML5)
Although it is clear that conformational changes in membrane proteins must occur for activity to be expressed, the exact structural rearrangements of these proteins has been difficult to determine given the lack of structural data for membrane proteins, especially as contrasted to water-soluble enzymes. Transporters allow slow (102 to 10-5 s-1) movement of ligands through pores which must be alternately open and closed to allow extracellular and intracellular access/egress. They need at a minimum an "open-to-out" and "open-to-in" conformational states. The pores must be transient and not continuous. Structural studies have been performed on E. Coli Lac Y Permease, a member of the largest transporter family, Major Facilitator Superfamily (MFS). These proteins have two halves with symmetrical halves each with 6 transmembrane domains. Crystal structures suggest a rocker switch like the figure above for the Glu transport protein.
Structural work by Singh et al on the Leu Transporter (LeuT), a member of the solute carrier 6 or sodium coupled transporters, which is an active transporter requiring movement of Na ion into the cells to power the uptake of Leu, show an "open-to-out" and occluded binding state for ligand (Leu). Tryptophan, a competitive nontransportable inhibitors binds to the open-to-out state, but is too large for the obligate occluded state so it is not transported.
Carriers of ligands need not be proteins. Ionophores are small organic molecules which can bind metal ions and move them down a concentration gradient across a bilayer. Most ionophores are not resident in the cell membrane. Rather they are mobile carriers of ions. They bind ions in solution, ferry them through the membrane, and then release them on the other side. As with receptor carrier proteins, they work in both directions but move ions in a net fashion down the concentration gradient of the ion. One example is valinomycin, a natural product of Streptomyces fulvissimus,. It is a cyclic peptide consisting of L and D-Val along with L-lactate and D-hydroxyisovalerate, connected through both ester and amide bonds.
![]()
![]() Jmol:
Valinomycin
Jmol:
Valinomycin
The structure of the valinomycin in its K+ form give clues to its function. The six Val carbonyl oxygens bind the K+ ion. The hydrophilic groups are pointed toward the center, while the hydrophobic groups point to the outside of the structure, allowing the K+ ion to be sequesters in a polar environment as the nonpolar exterior of the complex passes through the membrane. This ionophore is specific for K+ and binds the smaller Na+ ion weakly. This can be accounted for by two factors. The smaller sodium ion doesn't bind as tightly to the chelating carbonyl oxygens. Also, the sodium ion has a higher charge density, so the Na+/water interactions must be more stable and more difficult to break than those to K+. The ion must be desolvated before it binds to the complex. Other ionophores are specific for other ions.
Some ionophores, like gramicidin, from Bacillus brevis, forms a pore in the membrane through which different types of Group I ions may flow. It is a 15-residue peptide consisting of alternating D and L amino acids of significant hydrophobicity. The peptide appears to form a dimer helix (not an alpha helix, however), in a bilayer membrane.
![]()
![]() Jmol
: Gramicidin
Jmol
: Gramicidin
(go to the bottom link for next page in the top frame of this page and continue until you get to the Gramicidin page - an incredible Chime presentation.)
Aquaporins are tetrameric complexes that facilitate water diffusion through bilayers. They have amazing selectivity as they don't pass other solute or even protons (H3O+ or naked H+ ions). See the amazing link below to molecular dynamics simulations of the process.
![]()
![]() Jmol: Aquaporin
Jmol: Aquaporin
![]() Jmol: Updated
Aquaporin Monomer
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Aquaporin Monomer
Jmol14 (Java) |
JSMol (HTML5)
Gram negative bacteria (as well as mitochondria in eukaryotic cells) have protein complexes called porins (different from the aquaporin discussed above). The monomer porin forms a trimer in the membrane which forms a pore allowing small solute molecules necessary for bacterial cell growth to pass. The porin proteins share a 16 stranded anti-parallel beta barrel as a common motif. Solute molecules can pass through the pore created by the beta barrel. An example, maltoporin, is shown below:
![]() Jmol: Updated
Maltoporin Transport Protein
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Maltoporin Transport Protein
Jmol14 (Java) |
JSMol (HTML5)
Navigation
Return to Chapter 6A: Passive and Facilitated Diffusion Contents
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 6A: Passive and Facilitated Diffusion

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.